Abstract
Early T-Cell Precursor Acute Lymphoblastic Leukemia (ETP-ALL) is a treatment resistant type of pediatric leukemia whose biology is poorly understood, and new therapies are badly needed. While there are ongoing efforts to profile ETP-ALL using bulk and targeted genomics methods, these technologies fail to capture sub-clonal patterns in ETP-ALL, which may be pertinent to understanding its biology.
To synergize with current efforts to use bulk-genomics to characterize ETP-ALL, we performed single cell transcriptomics (scRNA-seq) and single cell chromatin accessibility (scATAC-seq) profiling on 30 ETP-ALL patients: 10 of which had excellent response to induction therapy (Day 29 minimal residual disease (MRD) <0.1%), 10 of which had poor response to induction therapy (Day 29 MRD >20%), and 10 of which responded to initial therapy but relapsed later. As additional comparator groups, we also profiled 10 typical T-ALL patients, and 10 Mixed Phenotype Acute Leukemia (MPAL) patients.
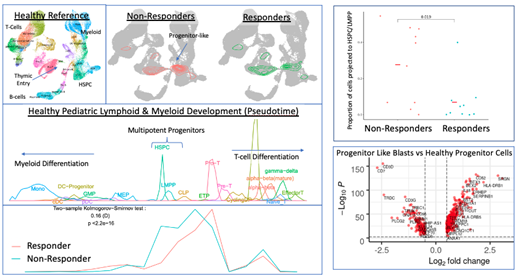
We mapped leukemic cells back to a healthy transcriptomic and epigenetic references, which we assembled using scRNA-seq and scATAC-seq of healthy pediatric bone marrow and thymus. We found that leukemic cells from different subtypes of ALL projected to different stages in healthy hematopoietic development. Non-ETP T-ALL patients had the highest proportions of cells projecting to more mature T-cell states, such as ab T cells, and MPAL patients had the highest proportions of cells projecting to myeloid progenitor cells (GMPs). However, the dominant projected population of ETP-ALL and MPAL were both lymphoid (CLP/LMPP) progenitors, supporting a high degree of overlap between these two treatment resistant cancers. Despite clear overall patterns, we found that there was both a large amount of inter-tumoral and intra-tumoral heterogeneity, with leukemic blasts from different patients and different populations within any one patient projecting to different cell states.
We then asked if any cell types were enriched in ETP-ALL patients that responded poorly to induction therapy (Day 29 MRD > 20%). We found that non-responders have a higher proportion of LMPP/HSPC (multi-potent) progenitor-like cells and that progenitor-like populations are transcriptomically and epigenetically different between non-responders and responders, with 100+ differentially expressed genes (DEGs) and 50+ differentially accessible chromatin regions. DEGs and enriched chromatin regions are enriched for transcription factor motifs known to regulate hematopoietic differentiation, such as BCL11A/B, MEF2A/B/C (non-responders enriched), TCF7, IRF9 (responders enriched).
We established patient derived xenograft (PDX) models for the patients in our dataset (28 of 50 have successfully engrafted thus far) by injection of primary blasts into immunodeficient (NSG) mice. We profiled 12 PDX models using scRNA-seq and found that blasts have a subtle, patient-specific transcriptomic shift upon engraftment, with 25-150 differentially expressed genes between patient and PDX. Interestingly, we found that cells in S/G2M cell cycle phase from PDX and patient are similar, while G1 cells have the largest transcriptomic shift, suggesting that a pool of cycling cells engrafts before settling in a subtly shifted transcriptomic state. Despite these observations, we found that the overall transcriptomic hierarchy is maintained in PDX, with engrafted cells from 5/6 PDX projecting to near-identical stages of arrest along our healthy hematopoietic trajectory, with preserved expression of key differentially expressed genes and transcription factors between non-responders and responders.
Our single cell multi-omics analysis reveals a large degree of intra and inter-tumoral transcriptomic and epigenetic heterogeneity in T-ALL, in which non-responding ETP-ALL patients have a higher proportion of blasts arrested in a HSPC/LMPP-like state. On-going mechanistic analyses are being performed using PDX models to assess the role of the progenitor-like cell populations in resistant disease and these analyses will be presented at ASH.
*CC and JX, as well as, DTT and KT contributed equally to this work
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal